In-house software
In-house software
03/09/17 10:34
A number of useful in-house programs have been produced that are available for download. Programs are either in C++ (making frequent use of the GSL), python (2.7, usually requiring numpy, scipy, matplotlib and nmrGlue modules) or a mixture of both. Outputs are produced in either gnuplot, matplotlib or latex. For programs with graphical user interfaces, wx-python will be required. Specific installation instructions and requirements are described for each program individually.
All have had testing on mac, linux and windows, on both 32 and 64bit chipsets.
Please get in touch if you have any problems with installing and using these guys!
[PLEASE NOTE: Download links are not setup yet! work in progress...]
All have had testing on mac, linux and windows, on both 32 and 64bit chipsets.
Please get in touch if you have any problems with installing and using these guys!
[PLEASE NOTE: Download links are not setup yet! work in progress...]
1. Sign determination by R1ρ
06/09/16 08:28
Python code to help with the determination of signs of chemical shift differences using the R1ρ method. The code takes a pb, kex and a list of chemical shift differences, and determines optimal B1 fields and offsets from numerically evolving the appropriate Louvillian. Effects of differences in transverse relaxation between exciting both upfield and downfield of the resonance of interest are calculated. Varian input macros files are produced, so that you only need type ‘go’ at the spectrometer. The program uses nmrPipe and nmrGlue to process data and will also analyze everything for you. Signs made simple!
Measurement of the signs of methyl chemical shift differences between ground and excited protein states by R1ρ: an application to αB-crystallin
Baldwin AJ, Kay LE
J. Biol. NMR (2012) 53(1) 1 pdf
Measurement of the signs of methyl chemical shift differences between ground and excited protein states by R1ρ: an application to αB-crystallin
Baldwin AJ, Kay LE
J. Biol. NMR (2012) 53(1) 1 pdf
2. Polymake
06/09/00 07:29
Python code for taking a pdb ‘building block’ file and assembling it into a polyhedra. Modelling restraints, such as enforcing a minimum distances between two points are implemented. Modified fortran code is provided for rapidly calculating PA collision cross section values, to enable models to be compared to experimental collision cross section data.
The polydispersity of αB-crystallin is rationalised by an interconverting polyhedral architecture
Baldwin AJ, Lioe H, Hilton GR, Baker LA, Rubinstein JL, Kay LE, Benesch JLP
Structure (2011), 19, 1855-63
pdf
supp
The polydispersity of αB-crystallin is rationalised by an interconverting polyhedral architecture
Baldwin AJ, Lioe H, Hilton GR, Baker LA, Rubinstein JL, Kay LE, Benesch JLP
Structure (2011), 19, 1855-63
supp
3. Champ
06/09/00 05:30
Used to analyze 1D electrospray mass spectra where a protein M is binding a ligand P giving complexes with stoichiometries of the form MiPj. The program assumes that the concentrations of the various complexes can be described by a continuous distribution function. We applied this approach in studies of clients binding to sHSP oligomers.
Dissecting heterogeneous molecular chaperone complexes using a mass spectrum deconvolution approach
Stengel F, Baldwin AJ, Bush MF, Hilton GR, Lioe H, Basha E, Jaya N, Vierling E, Benesch JLP
Chem & Biol. (2012), 19, 599-607
pdf
supp
commentary Slingsby C, Clark AR
Dissecting heterogeneous molecular chaperone complexes using a mass spectrum deconvolution approach
Stengel F, Baldwin AJ, Bush MF, Hilton GR, Lioe H, Basha E, Jaya N, Vierling E, Benesch JLP
Chem & Biol. (2012), 19, 599-607
supp
commentary Slingsby C, Clark AR
5. CPMG R2eff calculation
15/06/00 22:32
Implementations of various methods for calculating effective transverse relaxation rate in CPMG experiments for 2 site exchange. Includes exact solution to 2x2 Bloch McConnell equations (see publication 24).
c code
python code
An exact solution for R2,eff in CPMG experiments in the case of two site chemical exchange
Baldwin AJ
JMR (2014), 244, 114-124
pdf
supp
c code
python code
An exact solution for R2,eff in CPMG experiments in the case of two site chemical exchange
Baldwin AJ
JMR (2014), 244, 114-124
supp
6. IMPACT
03/07/99 14:46
Software for calculating the collision cross-section of proteins, for use in ion-mobility mass spectrometry calculations.
http://impact.chem.ox.ac.uk/
Collision Cross Sections for Structural Proteomics
Markland EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JLP
Structure (2015) 23(4):791-9
pdf
http://impact.chem.ox.ac.uk/
Collision Cross Sections for Structural Proteomics
Markland EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JLP
Structure (2015) 23(4):791-9
7. UNIDEC
03/06/99 14:47
Software for deconvolving mass spectra.
http://unidec.chem.ox.ac.uk/
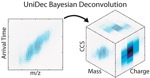
Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles
Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV
Anal. Chem. (2015) 87 (8) 4370-4376
pdf
http://unidec.chem.ox.ac.uk/

Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles
Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV
Anal. Chem. (2015) 87 (8) 4370-4376
8. MAGMA
03/05/99 14:47
Software for assigning methyl-methyl through space data, for use with a crystal structure using graph theory.
http://magma.chem.ox.ac.uk/
Automatic assignment of methyl-NMR spectra of supra-molecular machines using graph theory
Pritisanac I, Degiacomi M, Alderson R, Carneiro M, Eiso AB, Siegal G, Baldwin AJ,
J. Am. Chem. Soc. (in press) 2017
http://magma.chem.ox.ac.uk/
Automatic assignment of methyl-NMR spectra of supra-molecular machines using graph theory
Pritisanac I, Degiacomi M, Alderson R, Carneiro M, Eiso AB, Siegal G, Baldwin AJ,
J. Am. Chem. Soc. (in press) 2017
9. PULSAR
03/03/99 16:54
Pulsar, software for fitting gas phase dissociation data.
http://pulsar.chem.ox.ac.uk/
Quantifying the stabilizing effects of protein-ligand interactions in the gas phase
Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV
Nature Communications (2015) DOI: 10.1038/ncomms9551
pdf
http://pulsar.chem.ox.ac.uk/
Quantifying the stabilizing effects of protein-ligand interactions in the gas phase
Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV
Nature Communications (2015) DOI: 10.1038/ncomms9551